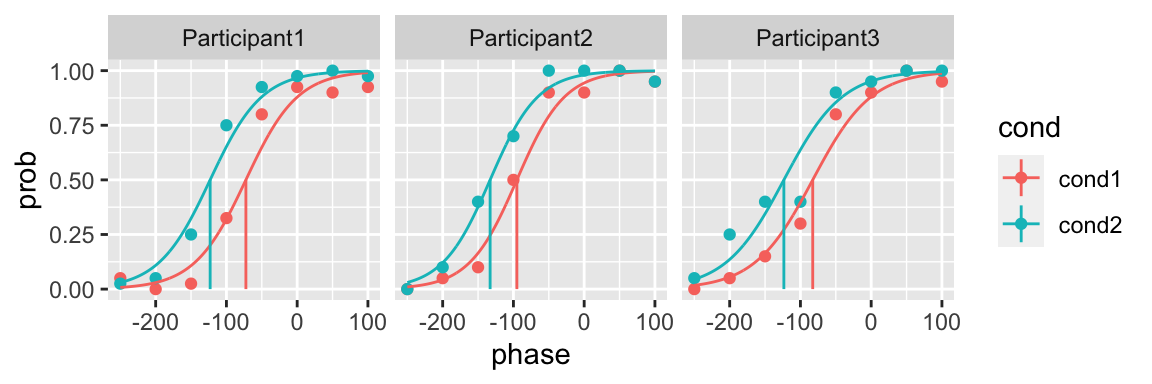
initial_parameters
Without providing initial parameters
If the function to fit (fun argument) is one of the predefined functions (cum_normal_fun, logistic_fun or weibull_fun), quickpsy calculates the initials parameters for you
library(quickpsy)
fit <- quickpsy(qpdat, phase, resp, grouping = c("participant", "cond"),
fun = logistic_fun,
bootstrap = "none")
plot(fit)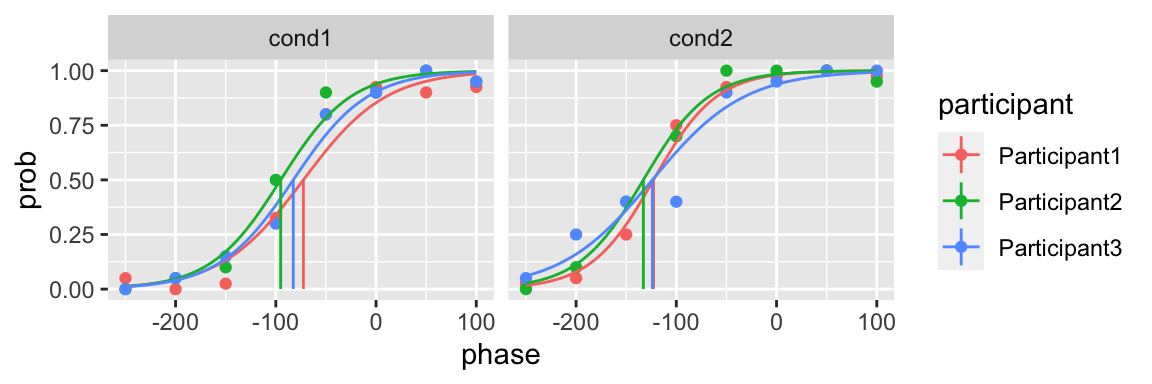
A vector initial parameters
The initial parameters could be a vector
library(quickpsy)
fit <- quickpsy(qpdat, phase, resp, grouping = c("participant", "cond"),
parini = c(-100, 70), # the default function is cum_normal_fun
bootstrap = "none")
plot(fit)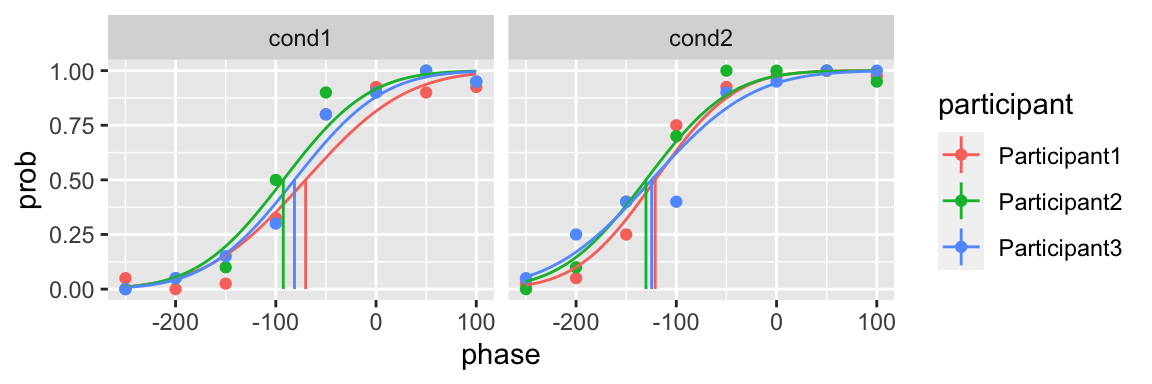
A list of initial parameters
The initial parameters could be a list in which each component constraints the lower and upper bounds of the parameter (for example, list(c(par1min, par1max), c(par2min, par2max)))
library(quickpsy)
fit <- quickpsy(qpdat, phase, resp, grouping = c("participant", "cond"),
parini = list(c(-300, 300), c(10, 200)),
bootstrap = "none")
plot(fit)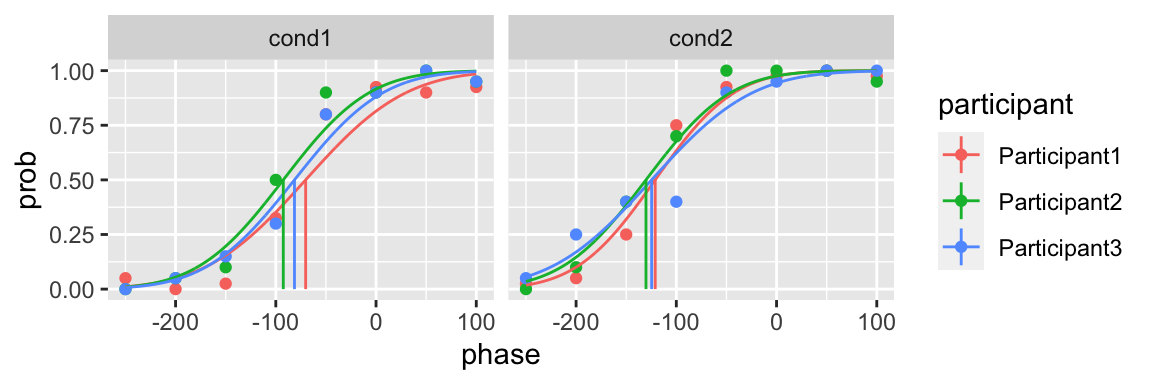 In addition of the upper and lower bound, you can also specify the initial guess of the parameters
In addition of the upper and lower bound, you can also specify the initial guess of the parameters
library(quickpsy)
fit <- quickpsy(qpdat, phase, resp, grouping = c("participant", "cond"),
parini = list(c(-300, 300), c(10, 200)),
parinivector = c(-100, 70),
bootstrap = "none")
plot(fit)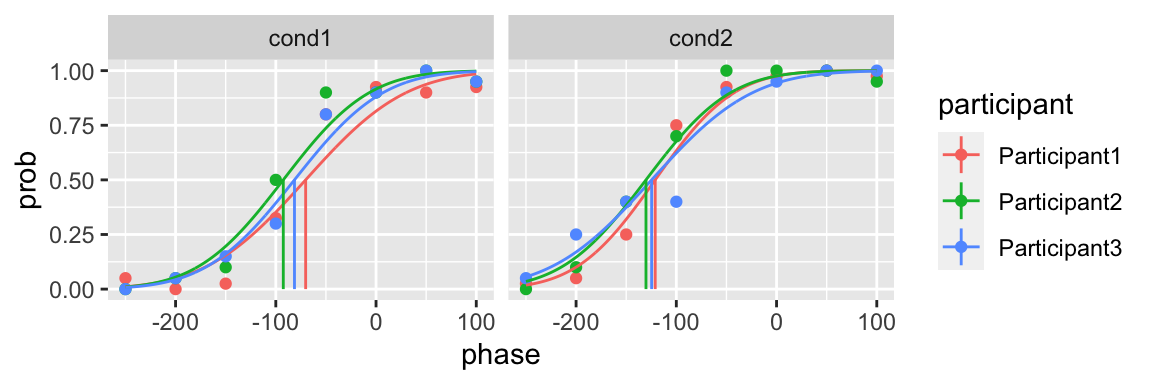
A dataframe of initial parameters
The initial parameters could be a dataframe specifiying the initial parameters for each condition. It should have the same structure that the component par of quickpsy. It is up to you how to build this dataframe. For example:
library(dplyr)
library(tidyr)
participants <- qpdat %>% distinct(participant)
conditions <- qpdat %>% distinct(cond)
parini <- conditions %>%
crossing(participants) %>%
crossing(tibble(parn = c("p1", "p2"))) %>%
arrange(cond) %>%
bind_cols(tibble(par = c(rep(c(-100, 70), 3),
rep(c(-150, 100), 3))))
parini
#> # A tibble: 12 x 4
#> cond participant parn par
#> <chr> <chr> <chr> <dbl>
#> 1 cond1 Participant1 p1 -100
#> 2 cond1 Participant1 p2 70
#> 3 cond1 Participant2 p1 -100
#> 4 cond1 Participant2 p2 70
#> 5 cond1 Participant3 p1 -100
#> 6 cond1 Participant3 p2 70
#> 7 cond2 Participant1 p1 -150
#> 8 cond2 Participant1 p2 100
#> 9 cond2 Participant2 p1 -150
#> 10 cond2 Participant2 p2 100
#> 11 cond2 Participant3 p1 -150
#> 12 cond2 Participant3 p2 100fit <- quickpsy(qpdat, phase, resp, grouping = c("participant", "cond"),
parini = parini, bootstrap = "none")
plot(fit) 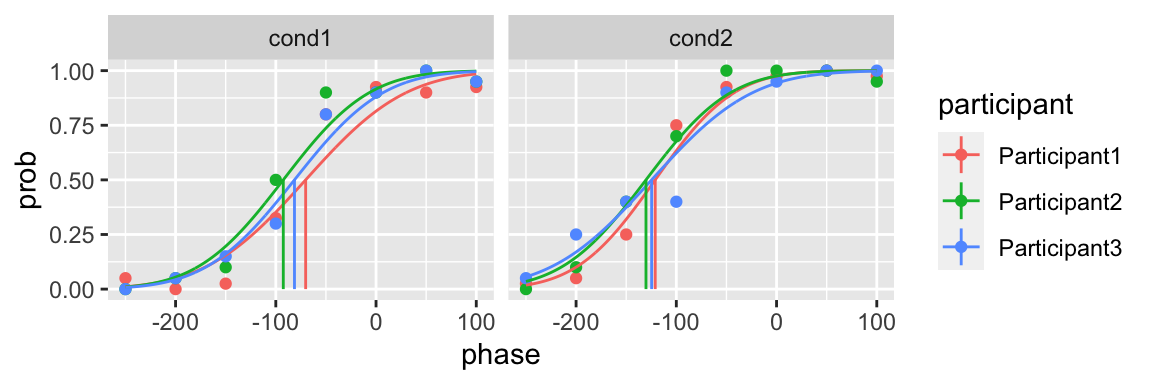
The dataframe could also contain the constraints of the parameters
parini <- conditions %>%
crossing(participants) %>%
crossing(tibble(parn = c("p1", "p2"))) %>%
arrange(cond) %>%
bind_cols(tibble(parmin = c(rep(c(-300, 10), 3),
rep(c(-500, 5), 3)),
parmax = c(rep(c(300, 200), 3),
rep(c(500, 400), 3))))
parini
#> # A tibble: 12 x 5
#> cond participant parn parmin parmax
#> <chr> <chr> <chr> <dbl> <dbl>
#> 1 cond1 Participant1 p1 -300 300
#> 2 cond1 Participant1 p2 10 200
#> 3 cond1 Participant2 p1 -300 300
#> 4 cond1 Participant2 p2 10 200
#> 5 cond1 Participant3 p1 -300 300
#> 6 cond1 Participant3 p2 10 200
#> 7 cond2 Participant1 p1 -500 500
#> 8 cond2 Participant1 p2 5 400
#> 9 cond2 Participant2 p1 -500 500
#> 10 cond2 Participant2 p2 5 400
#> 11 cond2 Participant3 p1 -500 500
#> 12 cond2 Participant3 p2 5 400Data frame of functions (dev version of quickpsy)
When the fun argument is a data frame of functions, you should specify all the parameters included in the functions
library(dplyr)
fun_df <- tibble(cond = c("cond1", "cond2"),
fun = c(function(x, p) (1 + exp(-p[2] * (x - p[1])))^(-1),
function(x, p) (1 + exp(-p[2] * (x - p[3])))^(-1)))
fit <- quickpsy(qpdat, phase, resp, grouping = c("participant", "cond"),
fun = fun_df, # shared slope parameter
parini = c(-100, 0.02, -100),
bootstrap = "none")
plot(fit, color = cond)