Kruskal-Wallis
The Kruskal-Wallis test uses the same logic that ANOVA, but instead of using the values of the data, it is using their ranks.
We want to study the effect of 3 fertilizers in plant growth (example from wikipedia). So the factor fertilizer has 3 levels or groups.
library(tidyverse)
g1 <- c(6, 8, 4, 5, 3, 4) # growth under fertilizer 1
g2 <- c(8, 12, 9, 11, 6, 8) # growth under fertilizer 2
g3 <- c(13, 9, 11, 8, 7, 12) # growth under fertilizer 3
d1 <- tibble(response = g1, fertilizer ="g1")
d2 <- tibble(response = g2, fertilizer ="g2")
d3 <- tibble(response = g3, fertilizer ="g3")
dat <- d1 %>%
bind_rows(d2) %>%
bind_rows(d3) %>%
mutate(fertilizer = as.factor(fertilizer))
dat## # A tibble: 18 × 2
## response fertilizer
## <dbl> <fct>
## 1 6 g1
## 2 8 g1
## 3 4 g1
## 4 5 g1
## 5 3 g1
## 6 4 g1
## 7 8 g2
## 8 12 g2
## 9 9 g2
## 10 11 g2
## 11 6 g2
## 12 8 g2
## 13 13 g3
## 14 9 g3
## 15 11 g3
## 16 8 g3
## 17 7 g3
## 18 12 g3We plot the data
ggplot(dat, aes(x = fertilizer, y = response)) +
geom_point()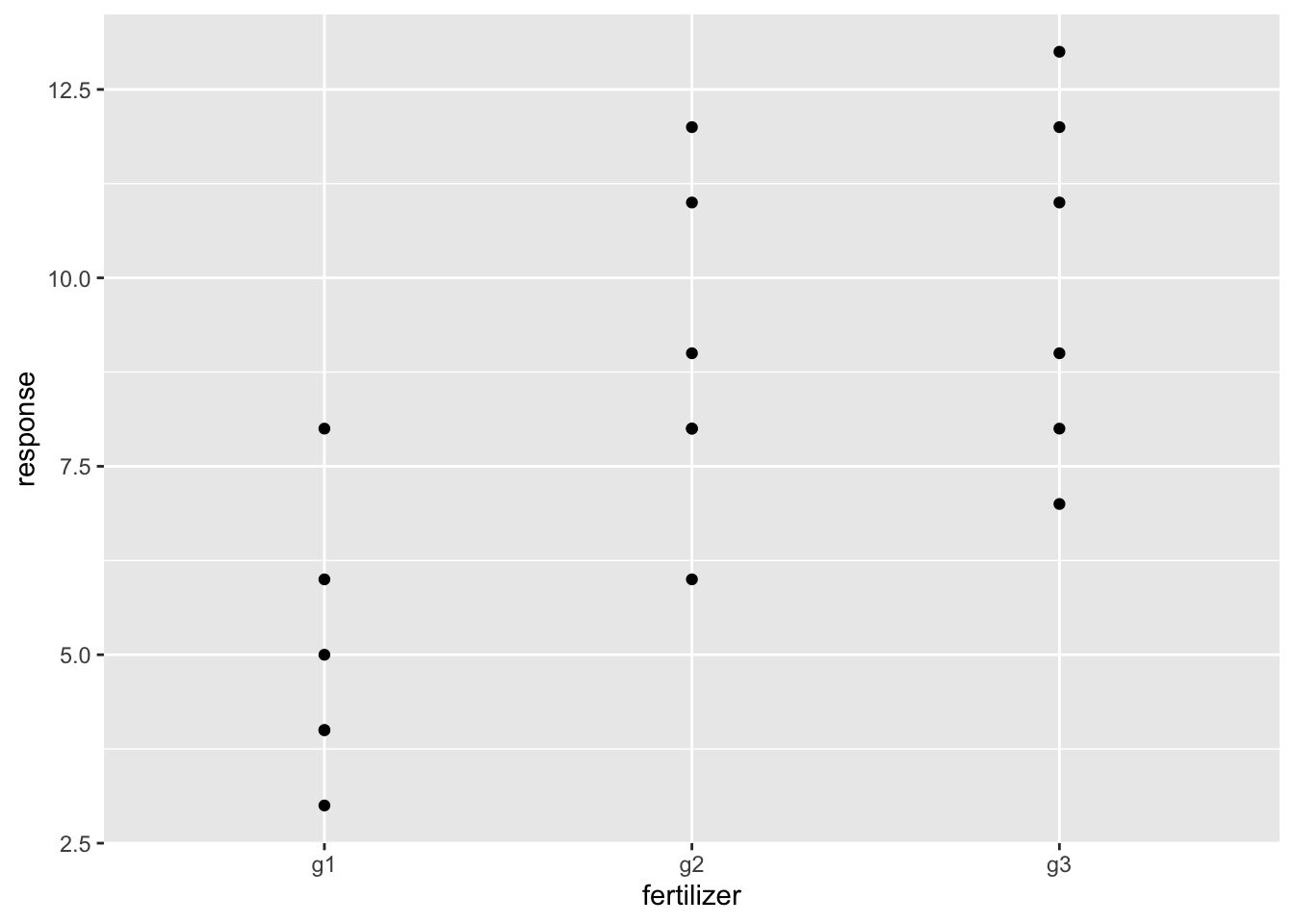
Running the test
kruskal.test(response ~ fertilizer, data = dat)##
## Kruskal-Wallis rank sum test
##
## data: response by fertilizer
## Kruskal-Wallis chi-squared = 9.4207, df = 2, p-value = 0.009002